Technology: Combining the Power of Epigenetics and the Consistency of qPCR
Since all cells in an individual have the same DNA blueprint, the functional distinction between different cell types is determined at the gene expression level. Epigenetic modification of DNA by methylation and demethylation is one of the mechanisms by which gene expression is controlled.
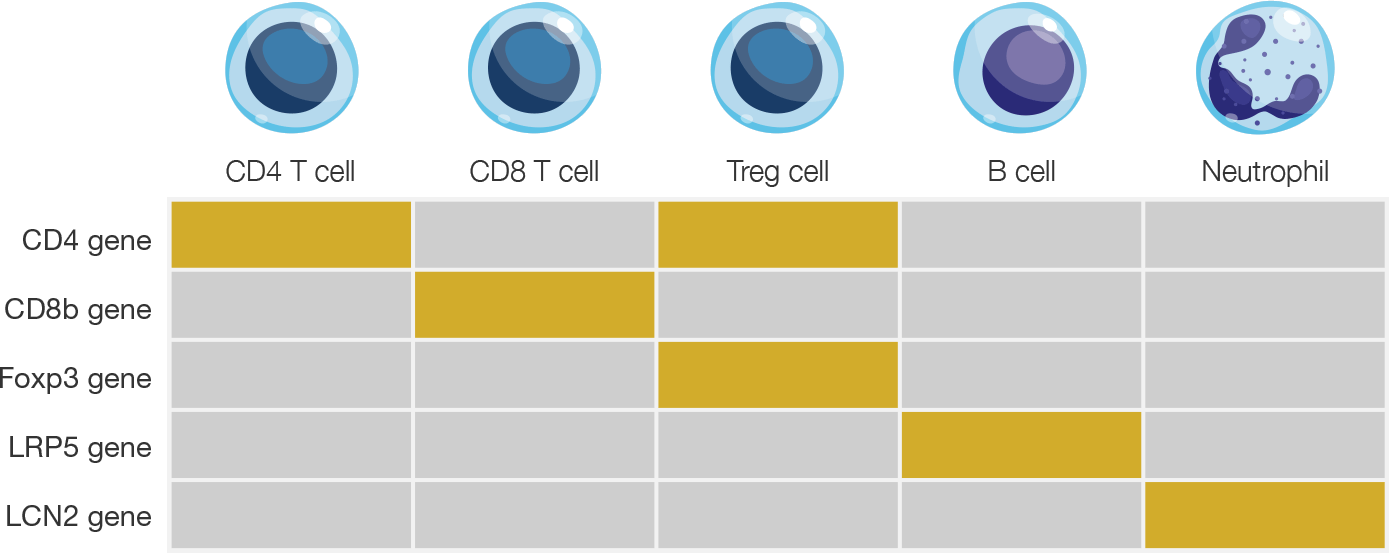
Epiontis ID harnesses the unique methylation patterns specific to distinct cells types, and uses a customized process and qPCR primers to amplify only those DNA regions that are demethylated in the cell type of interest. The quantitative nature of qPCR then allows for a precise count of the number of cells of interest in any sample.
Epigenetic methylation chart for select immune cell types
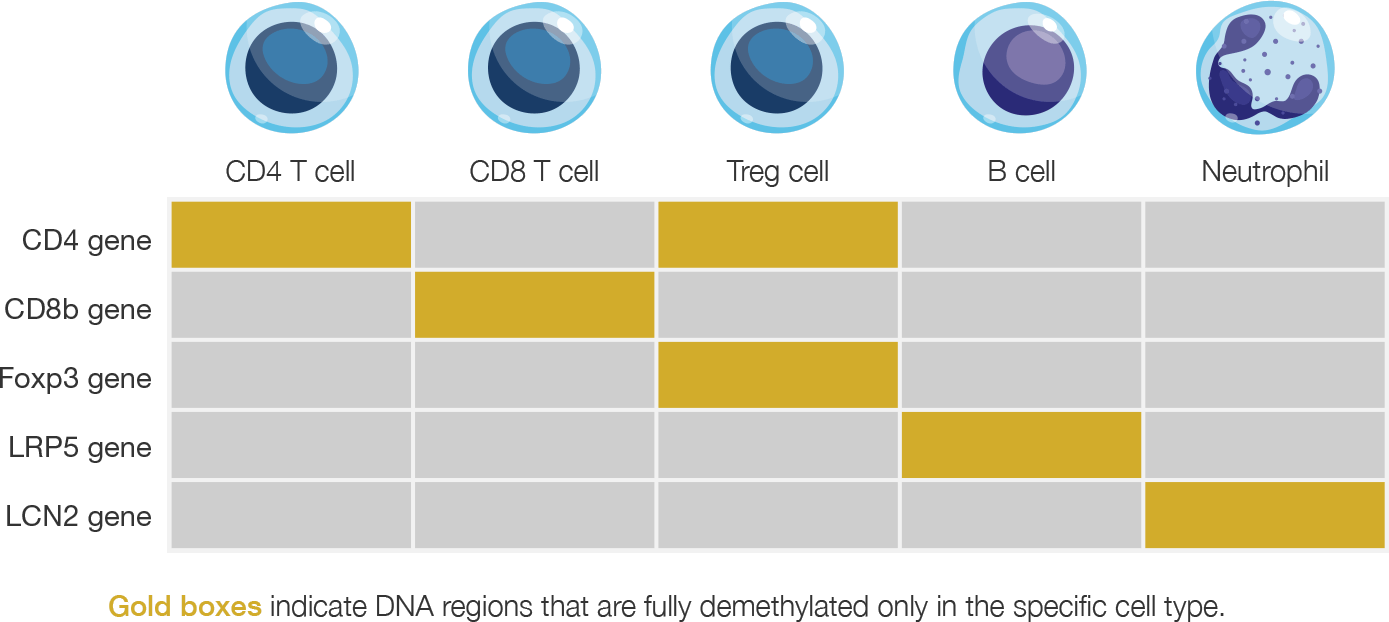
Gold boxes indicate DNA regions that are fully demethylated only in the specific cell type.
Consistent Results That Strongly Correlate to Flow Cytometry
Epiontis ID’s epigenetic-based cell counting has been reliably utilized for more than 10 years in more than 100 clinical studies.
The results generated by Epiontis ID are consistent across and within studies, and the data produced aligns with results from flow cytometry as shown in a study published in Science Translational Medicine.
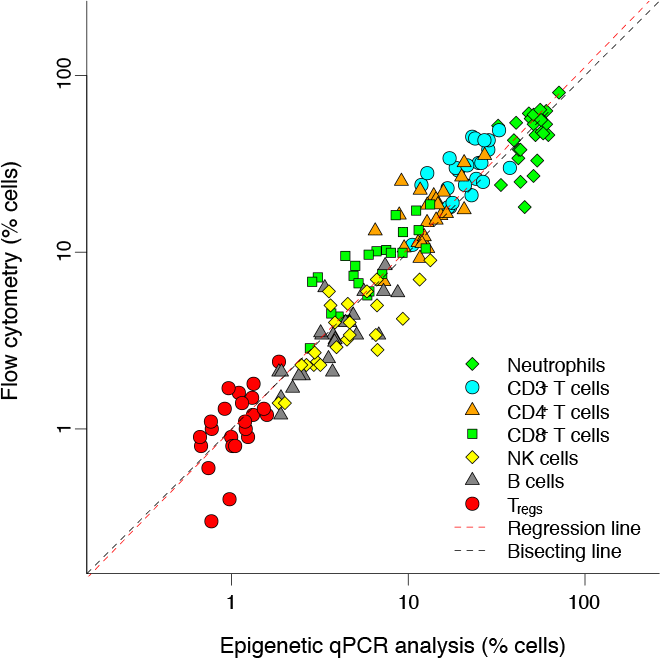
Comparison of Epiontis ID to flow cytometry in several cell types shows a high degree of correlation.
From Epigenetic immune cell counting in human blood samples for immunodiagnostics. Baron et al. Science Translational Medicine. 2018;10(452):eaan3508.
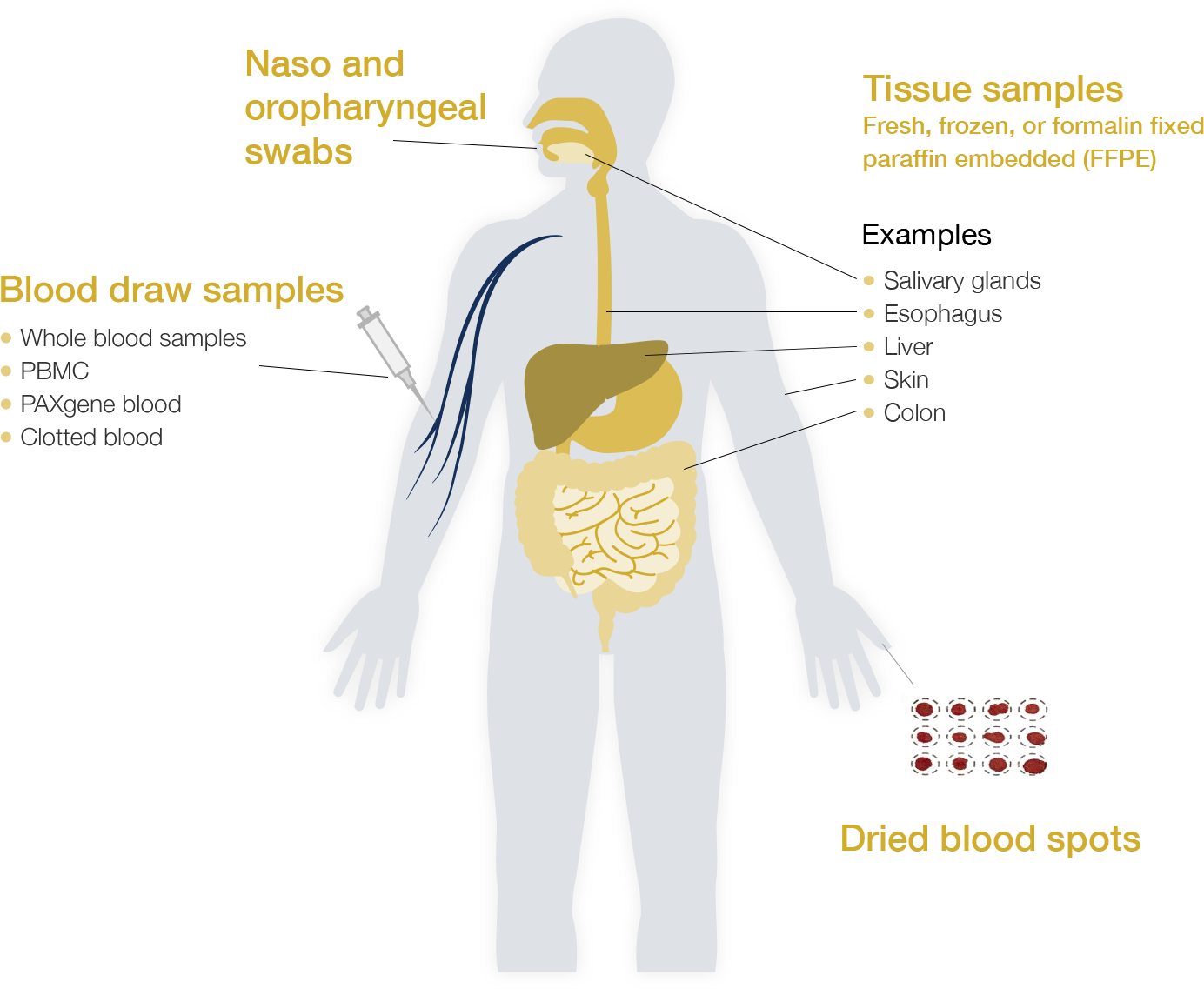
Broad Range of Sample Types Analyzed
Since Epiontis ID is a DNA-based methodology and epigenetic marks on DNA are highly stable, epigenetic immune-cell quantification methods can be applied to fresh, frozen, or paper-spotted dried blood and other body fluids or tissues, eliminating the need for special care during sample storage and transport. This creates opportunities for immune monitoring in studies where flow cytometry presents a logistical challenge.